MERFISH data analysis
Last updated: 2025-04-13
Checks: 6 1
Knit directory: KODAMA-Analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of
the R Markdown file created these results, you’ll want to first commit
it to the Git repo. If you’re still working on the analysis, you can
ignore this warning. When you’re finished, you can run
wflow_publish to commit the R Markdown file and build the
HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20240618) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 5f5ac63. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Untracked files:
Untracked: KODAMA.svg
Untracked: analysis/singlecell_datamatrix.Rmd
Untracked: analysis/singlecell_seurat.Rmd
Untracked: code/Acinar_Cell_Carcinoma.ipynb
Untracked: code/Adenocarcinoma.ipynb
Untracked: code/Adjacent_normal_section.ipynb
Untracked: code/DLFPC_preprocessing.R
Untracked: code/DLPFC - BANKSY.R
Untracked: code/DLPFC - BASS.R
Untracked: code/DLPFC - BAYESPACE.R
Untracked: code/DLPFC - Nonspatial.R
Untracked: code/DLPFC - PRECAST.R
Untracked: code/DLPFC_comparison.R
Untracked: code/DLPFC_results_analysis.R
Untracked: code/MERFISH - BANKSY.R
Untracked: code/MERFISH - BASS.R
Untracked: code/MERFISH - BAYESPACE.R
Untracked: code/MERFISH - Nonspatial.R
Untracked: code/MERFISH - PRECAST.R
Untracked: code/MERFISH_comparison.R
Untracked: code/MERFISH_results_analysis.R
Untracked: code/VisiumHD-CRC.ipynb
Untracked: code/VisiumHDassignment.py
Untracked: code/deep learning code DLPFC.R
Untracked: code/save tiles.py
Untracked: data/Adenocarcinoma.csv
Untracked: data/Annotations/
Untracked: data/DLFPC-Br5292-input.RData
Untracked: data/DLFPC-Br5595-input.RData
Untracked: data/DLFPC-Br8100-input.RData
Untracked: data/DLPFC-general.RData
Untracked: data/MERFISH-input.RData
Untracked: data/spots_classification_ALL.csv
Untracked: data/spots_classification_Acinar_Cell_Carcinoma.csv
Untracked: data/spots_classification_IF.csv
Untracked: data/spots_classification_Normal_prostate.csv
Untracked: data/trajectories.RData
Untracked: data/trajectories_VISIUMHD.RData
Untracked: output/BANSKY-results.RData
Untracked: output/BASS-results.RData
Untracked: output/BayesSpace-results.RData
Untracked: output/CRC-image.RData
Untracked: output/CRC-image2.RData
Untracked: output/CRC.png
Untracked: output/CRC2.png
Untracked: output/CRC7.png
Untracked: output/CRC8.png
Untracked: output/CRC_boxplot.png
Untracked: output/CRC_boxplot.svg
Untracked: output/CRC_boxplot2.svg
Untracked: output/CRC_linee.svg
Untracked: output/DL.RData
Untracked: output/DLFPC-All-2.RData
Untracked: output/DLFPC-All.RData
Untracked: output/DLFPC-Br5292.RData
Untracked: output/DLFPC-Br5595.RData
Untracked: output/DLFPC-Br8100.RData
Untracked: output/DLFPC-variablesXdeeplearning.RData
Untracked: output/DLPFC-BANSKY-results.RData
Untracked: output/DLPFC-BASS-results.RData
Untracked: output/DLPFC-BayesSpace-results.RData
Untracked: output/DLPFC-Nonspatial-results.RData
Untracked: output/DLPFC-PRECAST-results.RData
Untracked: output/DLPFC_all_cluster.svg
Untracked: output/DLPFCpathway.RData
Untracked: output/Figure 1 - boxplot.pdf
Untracked: output/Figure 2 - DLPFC 10.pdf
Untracked: output/Figures/
Untracked: output/KODAMA-results.RData
Untracked: output/KODAMA_DLPFC_All_original.svg
Untracked: output/KODAMA_DLPFC_Br5595.svg
Untracked: output/KODAMA_DLPFC_Br5595_slide.svg
Untracked: output/Loupe.csv
Untracked: output/MERFISH-BANSKY-results.RData
Untracked: output/MERFISH-BASS-results.RData
Untracked: output/MERFISH-BayesSpace-results.RData
Untracked: output/MERFISH-KODAMA-results.RData
Untracked: output/MERFISH-Nonspatial-results.RData
Untracked: output/MERFISH-PRECAST-results.RData
Untracked: output/MERFISH.RData
Untracked: output/Nonspatial-results.RData
Untracked: output/Prostate.RData
Untracked: output/VisiumHD-RNA.RData
Untracked: output/VisiumHD-genes.pdf
Untracked: output/VisiumHD.RData
Untracked: output/boh.svg
Untracked: output/desmoplastic_distance_carcinoma.csv
Untracked: output/image.RData
Untracked: output/pp.RData
Untracked: output/pp2.RData
Untracked: output/pp3.RData
Untracked: output/pp4.RData
Untracked: output/pp5.RData
Untracked: output/prostate1.svg
Untracked: output/prostate2.svg
Untracked: output/prostate3.svg
Untracked: output/prostate4.svg
Untracked: output/prostate5.svg
Untracked: output/prostate6.svg
Untracked: output/prostate7.svg
Untracked: output/subclusters1.csv
Untracked: output/subclusters2.csv
Untracked: output/subclusters3.csv
Untracked: output/tight_boundary.geojson
Untracked: output/trajectory.csv
Unstaged changes:
Deleted: analysis/D1.Rmd
Deleted: analysis/DLPFC-12.Rmd
Deleted: analysis/DLPFC-4.Rmd
Modified: analysis/DLPFC.Rmd
Deleted: analysis/DLPFC1.Rmd
Deleted: analysis/DLPFC10.Rmd
Deleted: analysis/DLPFC2.Rmd
Deleted: analysis/DLPFC3.Rmd
Deleted: analysis/DLPFC4.Rmd
Deleted: analysis/DLPFC5.Rmd
Deleted: analysis/DLPFC6.Rmd
Deleted: analysis/DLPFC7.Rmd
Deleted: analysis/DLPFC8.Rmd
Deleted: analysis/DLPFC9.Rmd
Deleted: analysis/Du1.Rmd
Deleted: analysis/Du10.Rmd
Deleted: analysis/Du11.Rmd
Deleted: analysis/Du12.Rmd
Deleted: analysis/Du13.Rmd
Deleted: analysis/Du14.Rmd
Deleted: analysis/Du15.Rmd
Deleted: analysis/Du16.Rmd
Deleted: analysis/Du17.Rmd
Deleted: analysis/Du18.Rmd
Deleted: analysis/Du19.Rmd
Deleted: analysis/Du2.Rmd
Deleted: analysis/Du20.Rmd
Deleted: analysis/Du3.Rmd
Deleted: analysis/Du4.Rmd
Deleted: analysis/Du5.Rmd
Deleted: analysis/Du6.Rmd
Deleted: analysis/Du7.Rmd
Deleted: analysis/Du8.Rmd
Deleted: analysis/Du9.Rmd
Modified: analysis/Giotto.Rmd
Modified: analysis/MERFISH.Rmd
Deleted: analysis/MERFISH1a (copy).Rmd
Deleted: analysis/MERFISH1a.Rmd
Deleted: analysis/MERFISH1b (copy).Rmd
Deleted: analysis/MERFISH1b.Rmd
Deleted: analysis/MERFISH2a (copy).Rmd
Deleted: analysis/MERFISH2a.Rmd
Deleted: analysis/MERFISH2b (copy).Rmd
Deleted: analysis/MERFISH2b.Rmd
Deleted: analysis/MERFISH3a (copy).Rmd
Deleted: analysis/MERFISH3a.Rmd
Deleted: analysis/MERFISH3b (copy).Rmd
Deleted: analysis/MERFISH3b.Rmd
Deleted: analysis/MERFISH4a (copy).Rmd
Deleted: analysis/MERFISH4a.Rmd
Deleted: analysis/MERFISH4b (copy).Rmd
Deleted: analysis/MERFISH4b.Rmd
Modified: analysis/Prostate.Rmd
Deleted: analysis/STARmap.Rmd
Modified: analysis/Seurat.Rmd
Deleted: analysis/Simulation.Rmd
Deleted: analysis/Single-cell.Rmd
Modified: analysis/SpatialExperiment.Rmd
Modified: analysis/VisiumHD.Rmd
Modified: code/VisiumHD_CRC_download.sh
Deleted: data/Pathology.csv
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/MERFISH.Rmd) and HTML
(docs/MERFISH.html) files. If you’ve configured a remote
Git repository (see ?wflow_git_remote), click on the
hyperlinks in the table below to view the files as they were in that
past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 74c3d1e | Stefano Cacciatore | 2025-01-10 | Start my new project |
| html | 432fc49 | Stefano Cacciatore | 2025-01-10 | Build site. |
| Rmd | 52e75ec | Stefano Cacciatore | 2025-01-10 | Start my new project |
| html | 2c6734f | Stefano Cacciatore | 2024-09-05 | Build site. |
| Rmd | 6565005 | Stefano Cacciatore | 2024-09-05 | Start my new project |
| html | d1192e9 | Stefano Cacciatore | 2024-08-12 | Build site. |
| html | 3374e66 | Stefano Cacciatore | 2024-08-06 | Build site. |
| Rmd | d5e7c3c | Stefano Cacciatore | 2024-08-05 | Start my new project |
| html | 35ce733 | Stefano Cacciatore | 2024-08-03 | Build site. |
| html | 82fe167 | Stefano Cacciatore | 2024-07-24 | Build site. |
| html | 6f7daac | Stefano Cacciatore | 2024-07-19 | Build site. |
| html | 7be8f59 | tkcaccia | 2024-07-15 | updates |
| Rmd | f8ca54a | tkcaccia | 2024-07-14 | update |
| html | f8ca54a | tkcaccia | 2024-07-14 | update |
| Rmd | 0638f93 | GitHub | 2024-07-08 | Update MERFISH.Rmd |
| html | d3de9e1 | GitHub | 2024-07-08 | Update MERFISH.html |
| html | 32f206e | GitHub | 2024-07-08 | Update MERFISH.html |
| html | 940ec13 | GitHub | 2024-07-04 | Update MERFISH.html |
| Rmd | 9ee53df | GitHub | 2024-06-26 | Update MERFISH.Rmd |
| html | 35e8f25 | Stefano Cacciatore | 2024-06-26 | Update docs with MERFISH.html |
| Rmd | c3eee75 | GitHub | 2024-06-26 | Update MERFISH.Rmd |
| Rmd | e7ddec6 | GitHub | 2024-06-26 | Update MERFISH.Rmd |
| Rmd | eb29180 | GitHub | 2024-06-26 | Update MERFISH.Rmd |
| Rmd | bd1988e | GitHub | 2024-06-26 | Update MERFISH.Rmd |
| Rmd | 911564c | GitHub | 2024-06-26 | Update MERFISH.Rmd |
| Rmd | 246f0bd | GitHub | 2024-06-25 | Update MERFISH.Rmd |
| html | 20a6dac | Stefano Cacciatore | 2024-06-25 | Update docs with MERFISH.html |
| Rmd | e56c4ff | GitHub | 2024-06-20 | Update MERFISH.Rmd |
| Rmd | 9df203d | GitHub | 2024-06-20 | Update MERFISH.Rmd |
| Rmd | be3cdeb | GitHub | 2024-06-20 | Update MERFISH.Rmd |
| Rmd | 2e7fa5e | GitHub | 2024-06-20 | Update MERFISH.Rmd |
| Rmd | a21b903 | GitHub | 2024-06-20 | Update MERFISH.Rmd |
| html | ee4ee17 | GitHub | 2024-06-19 | Add files via upload |
| Rmd | 615fc05 | GitHub | 2024-06-19 | Add files via upload |
Introduction
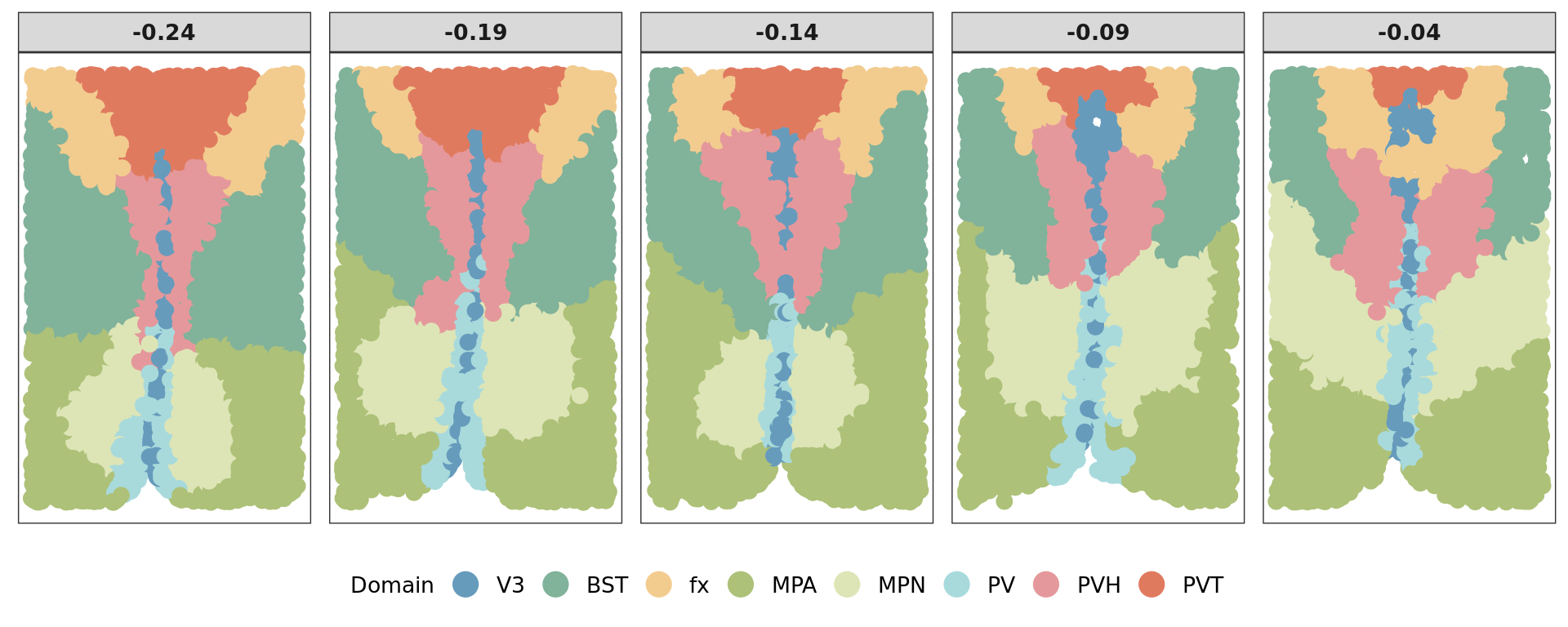
Here, we apply KODAMA to analyze the spatial transcriptomic data that measured the mouse preoptic region of the hypothalamus using the MERFISH technology from Moffitt et al., 2018. Link to study. We focus on the tissue sections Bregma -0.04, -0.09, -0.14, -0.19, and -0.24 mm from a consecutive brain hypothalamic region of animal 1. Data was retrieved from the BASS GitHub repository The original data can be downloaded from Dryad.
Loading and Preprocessing Data
The R library are loaded and the number of parallel computation is set.
# Load the necessary libraries
library(rgl)
library(irlba)
library(KODAMAextra)
library(scater)
library(SPARK)
library(ggplot2)
library(plotly)
library(mclust)
library(harmony)
library(bluster)
library(igraph)
n.cores=16The spatial ttranscriptomic data and relative spatial coordinates are loaded in the R environment.
# Load the MERFISH data
load("../MERFISH_Animal1.RData")
cols <- c("#669bbc", "#81b29a", "#f2cc8f", "#adc178",
"#dde5b6", "#a8dadc", "#e5989b", "#e07a5f",
"#aae5b6", "#a8aadc", "#e59811", "#aa7900")
# Define the slides to be analyzed
slides <- c("-0.04", "-0.09", "-0.14", "-0.19", "-0.24")
# Initialize variables
xyz <- NULL
tissue_segments <- NULL
cell_type <- NULL
RNA <- NULL
# Extract spatial and expression data from each slide
for (i in slides) {
x <- info_mult[[i]]$x / 1000
y <- info_mult[[i]]$y / 1000
z <- as.numeric(i)
slide_xyz <- cbind(x - min(x), y - min(y), z)
xyz <- rbind(xyz, slide_xyz)
tissue_segments <- c(tissue_segments, info_mult[[i]]$z)
cell_type <- c(cell_type, info_mult[[i]]$Cell_class)
RNA <- rbind(RNA, t(cnts_mult[[i]]))
}
# Normalize RNA counts
RNA <- t(normalizeCounts(t(RNA), log = TRUE))
# Convert tissue segments to factor with defined levels
tissue_segments <- factor(tissue_segments, levels = c("V3", "BST", "fx", "MPA", "MPN", "PV", "PVH", "PVT"))
# Convert xyz to numeric matrix
xyz <- matrix(as.numeric(as.matrix(xyz)), ncol = ncol(xyz))Identifying Differentially Expressed Genes
Genes are ranked using the SPARK-X algorithm independently for each slide. The top gene are the spatially expressed genes.
top=multi_SPARKX(RNA,xyz[,-3],as.factor(xyz[,3]),n.cores = n.cores)Passing message
The gene expression value are processed using the passing-message function.
RNA.PM=passing.message(RNA[,top[1:100]],xyz)Dimensionality reduction
The dimensionality of the dataset is reduced using the principal component analysis (PCA) and batch-effect is removed using the harmony function.
RNA.PM.scaled=scale(RNA.PM)
pca_results <- irlba(A = RNA.PM.scaled, nv = 50)
pca.PM <- pca_results$u %*% diag(pca_results$d)
pca.PM=RunHarmony(pca.PM,data.frame(z=xyz[,3]),"z")
RNA.scaled=scale(RNA[,top[1:100]])
pca_results <- irlba(A = RNA.scaled, nv = 50)
pca <- pca_results$u %*% diag(pca_results$d)
par(mfrow = c(1, 2))
plot(pca, col = cols[tissue_segments], main = "PCA",pch=20,xlab="PC1",ylab="PC2")
plot(pca.PM, col = cols[tissue_segments], main = "PCA.PM",pch=20,xlab="PC1",ylab="PC2")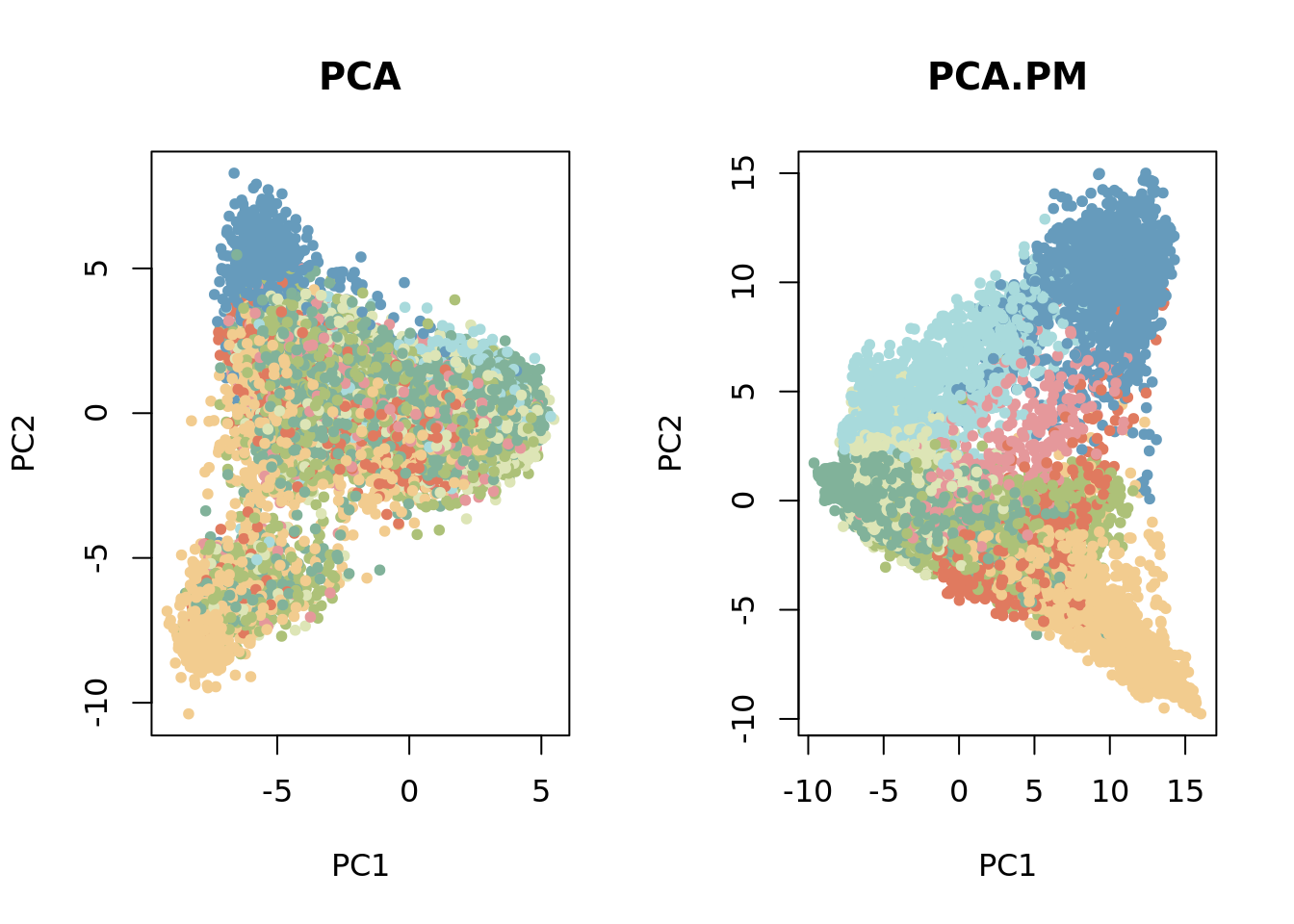
Applying KODAMA
KODAMA is performed of the first 20 principal components of PCA.
# Apply KODAMA to the PCA results
jj=KODAMA.matrix.parallel(pca.PM[,1:20],
spatial = xyz,
landmarks = 100000,
n.cores=n.cores,
seed = 543210)Calculating Network
Calculating Network spatialsocket cluster with 16 nodes on host 'localhost'
================================================================================
Finished parallel computation
[1] "Calculation of dissimilarity matrix..."
================================================================================config=umap.defaults
config$n_neighbors=30
config$n_threads = n.cores
vis <- KODAMA.visualization(jj, method = "UMAP",config=config)Clustering and Refinement
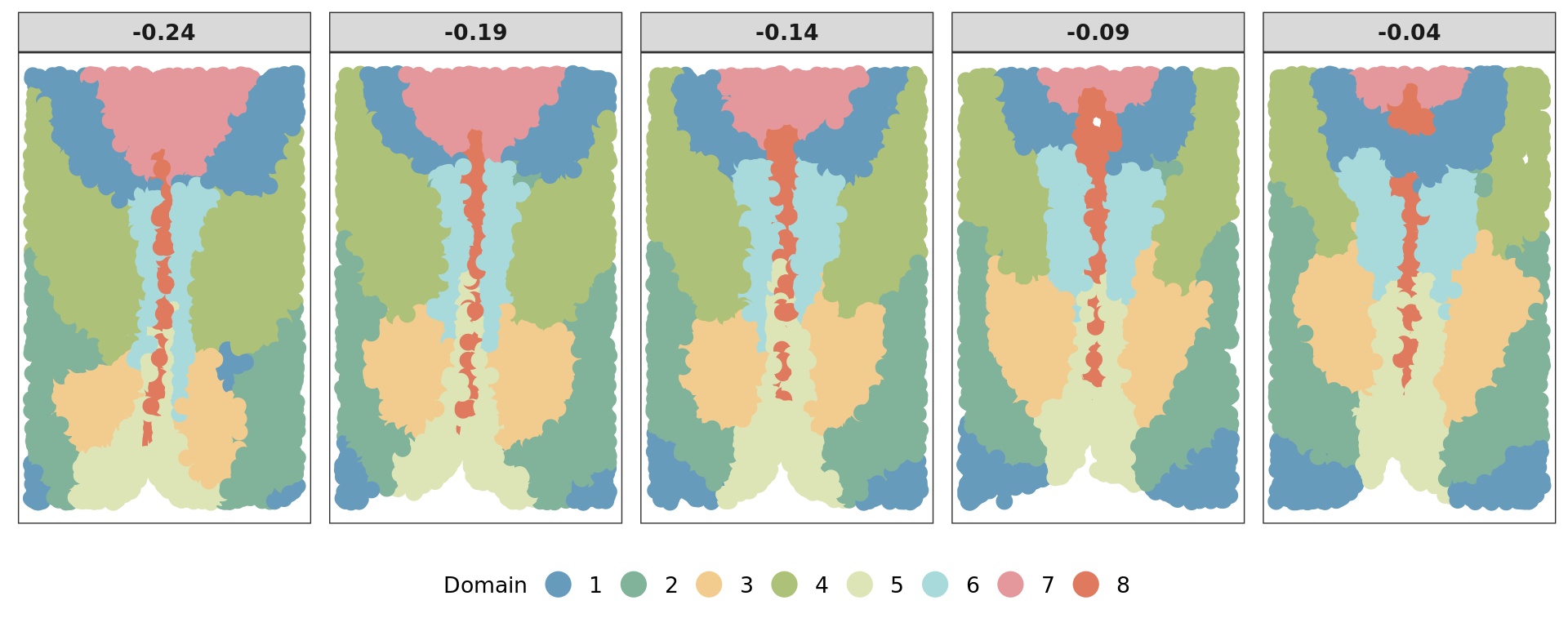
A louvain clustering is performed.
g <- makeSNNGraph(as.matrix(vis), k = 100)
clu= louvain(g,ncluster = 8)$membership[1] 0 1
[1] 0.2 8.0ref=refine_SVM(xyz,clu,cost=1000)[1] "1"[1] 0.4927165 0.6436110 0.5047492 0.6667729 0.6808519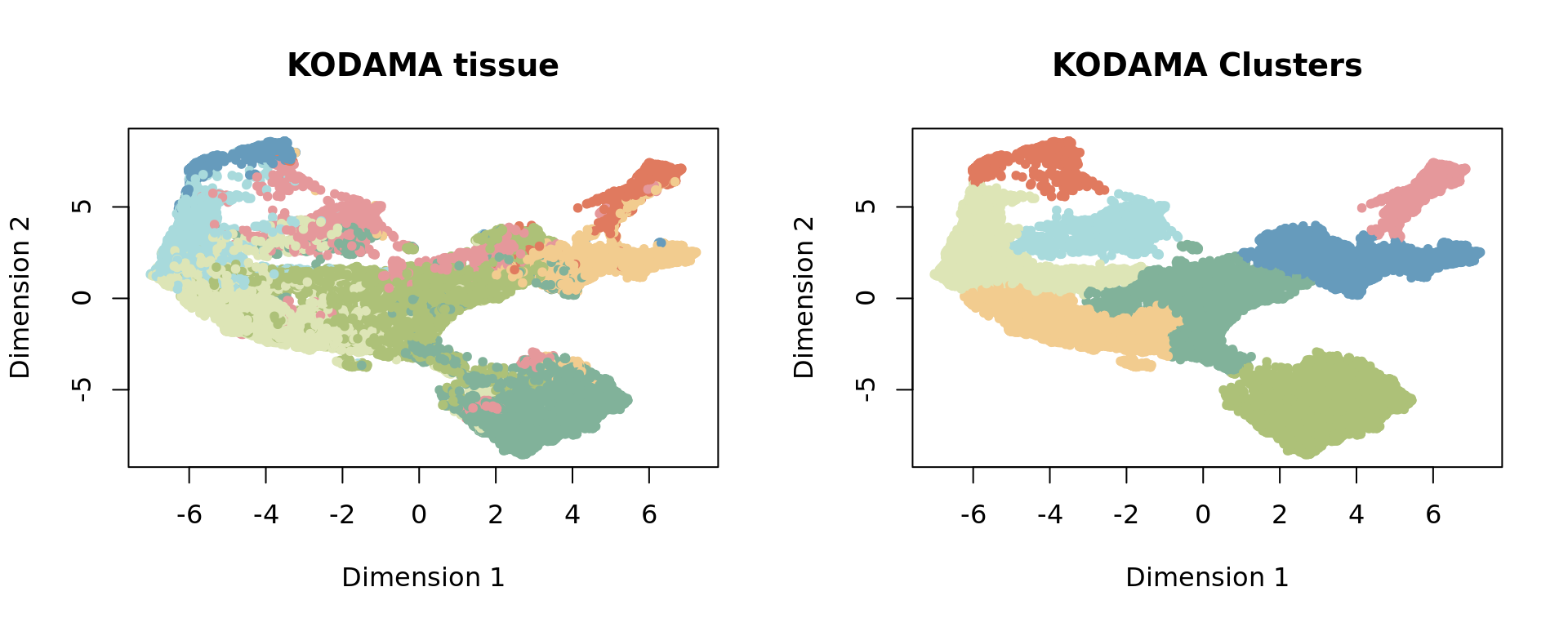
 # 3D Visualization of Slice -0.14
# 3D Visualization of Slice -0.14
library(rgl)
library(MASS)
library(misc3d)
volume_rendering(xyz[!is.na(ref),],ref[!is.na(ref)],selection = c("3","7","8","6"),alpha = c(0.8,0.8,0.8,0.8,0.8,0.8,0.8,0.8) ,colors = cols,cells=c(20, 20, 20),level=exp(2.4))
sessionInfo()R version 4.4.3 (2025-02-28)
Platform: x86_64-pc-linux-gnu
Running under: Ubuntu 20.04.6 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.9.0
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.9.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
time zone: Etc/UTC
tzcode source: system (glibc)
attached base packages:
[1] stats4 parallel stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] BayesSpace_1.14.0 Seurat_5.2.1
[3] SeuratObject_5.0.2 sp_2.2-0
[5] PRECAST_1.6.6 ProFAST_1.6
[7] gtools_3.9.5 SpatialExperiment_1.14.0
[9] Banksy_1.0.0 BASS_1.1.0.017
[11] GIGrvg_0.8 misc3d_0.9-1
[13] MASS_7.3-65 igraph_2.1.4
[15] bluster_1.14.0 harmony_1.2.3
[17] Rcpp_1.0.14 mclust_6.1.1
[19] plotly_4.10.4 SPARK_1.1.1
[21] scater_1.32.1 ggplot2_3.5.1
[23] scuttle_1.14.0 SingleCellExperiment_1.26.0
[25] SummarizedExperiment_1.34.0 Biobase_2.64.0
[27] GenomicRanges_1.56.2 GenomeInfoDb_1.40.1
[29] IRanges_2.38.1 S4Vectors_0.42.1
[31] BiocGenerics_0.50.0 MatrixGenerics_1.16.0
[33] matrixStats_1.5.0 KODAMAextra_1.2
[35] e1071_1.7-16 doParallel_1.0.17
[37] iterators_1.0.14 foreach_1.5.2
[39] KODAMA_3.0 umap_0.2.10.0
[41] Rtsne_0.17 minerva_1.5.10
[43] irlba_2.3.5.1 Matrix_1.7-3
[45] rgl_1.3.18 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] DirichletReg_0.7-1 goftest_1.2-3
[3] vctrs_0.6.5 spatstat.random_3.3-3
[5] digest_0.6.37 png_0.1-8
[7] proxy_0.4-27 git2r_0.33.0
[9] ggrepel_0.9.6 deldir_2.0-4
[11] parallelly_1.43.0 combinat_0.0-8
[13] Rnanoflann_0.0.3 magick_2.8.6
[15] reshape2_1.4.4 GiRaF_1.0.1
[17] httpuv_1.6.15 withr_3.0.2
[19] xfun_0.51 ggpubr_0.6.0
[21] survival_3.8-3 memoise_2.0.1
[23] ggbeeswarm_0.7.2 zoo_1.8-13
[25] pbapply_1.7-2 Formula_1.2-5
[27] promises_1.3.2 httr_1.4.7
[29] rstatix_0.7.2 rhdf5filters_1.16.0
[31] globals_0.16.3 fitdistrplus_1.2-2
[33] rhdf5_2.48.0 ps_1.9.0
[35] rstudioapi_0.17.1 UCSC.utils_1.0.0
[37] miniUI_0.1.1.1 generics_0.1.3
[39] base64enc_0.1-3 processx_3.8.6
[41] curl_6.2.2 zlibbioc_1.50.0
[43] ScaledMatrix_1.12.0 polyclip_1.10-7
[45] doSNOW_1.0.20 GenomeInfoDbData_1.2.12
[47] SparseArray_1.4.8 xtable_1.8-4
[49] stringr_1.5.1 pracma_2.4.4
[51] evaluate_1.0.3 S4Arrays_1.4.1
[53] BiocFileCache_2.12.0 colorspace_2.1-1
[55] filelock_1.0.3 ROCR_1.0-11
[57] reticulate_1.42.0 spatstat.data_3.1-6
[59] magrittr_2.0.3 lmtest_0.9-40
[61] later_1.4.1 viridis_0.6.5
[63] lattice_0.22-7 spatstat.geom_3.3-6
[65] future.apply_1.11.3 getPass_0.2-4
[67] scattermore_1.2 cowplot_1.1.3
[69] RcppAnnoy_0.0.22 class_7.3-23
[71] pillar_1.10.1 nlme_3.1-168
[73] compiler_4.4.3 beachmat_2.20.0
[75] RSpectra_0.16-2 stringi_1.8.7
[77] tensor_1.5 plyr_1.8.9
[79] crayon_1.5.3 abind_1.4-8
[81] locfit_1.5-9.12 bit_4.6.0
[83] sandwich_3.1-1 dplyr_1.1.4
[85] whisker_0.4.1 codetools_0.2-20
[87] BiocSingular_1.20.0 openssl_2.3.2
[89] bslib_0.9.0 DR.SC_3.5
[91] mime_0.13 splines_4.4.3
[93] fastDummies_1.7.5 dbplyr_2.5.0
[95] sparseMatrixStats_1.16.0 maxLik_1.5-2.1
[97] knitr_1.50 blob_1.2.4
[99] fs_1.6.5 listenv_0.9.1
[101] label.switching_1.8 DelayedMatrixStats_1.26.0
[103] ggsignif_0.6.4 RcppHungarian_0.3
[105] tibble_3.2.1 statmod_1.5.0
[107] callr_3.7.6 lpSolve_5.6.23
[109] pkgconfig_2.0.3 tools_4.4.3
[111] cachem_1.1.0 aricode_1.0.3
[113] RhpcBLASctl_0.23-42 RSQLite_2.3.9
[115] viridisLite_0.4.2 DBI_1.2.3
[117] fastmap_1.2.0 rmarkdown_2.29
[119] scales_1.3.0 grid_4.4.3
[121] ica_1.0-3 broom_1.0.8
[123] sass_0.4.9 coda_0.19-4.1
[125] patchwork_1.3.0 dotCall64_1.2
[127] carData_3.0-5 RANN_2.6.2
[129] snow_0.4-4 farver_2.1.2
[131] yaml_2.3.10 ggthemes_5.1.0
[133] cli_3.6.4 purrr_1.0.4
[135] lifecycle_1.0.4 dbscan_1.2.2
[137] askpass_1.2.1 uwot_0.2.3
[139] backports_1.5.0 BiocParallel_1.38.0
[141] gtable_0.3.6 rjson_0.2.23
[143] ggridges_0.5.6 progressr_0.15.1
[145] limma_3.60.6 edgeR_4.2.2
[147] jsonlite_2.0.0 miscTools_0.6-28
[149] RcppHNSW_0.6.0 bitops_1.0-9
[151] xgboost_1.7.9.1 bit64_4.6.0-1
[153] assertthat_0.2.1 spatstat.utils_3.1-3
[155] BiocNeighbors_1.22.0 matlab_1.0.4.1
[157] metapod_1.12.0 jquerylib_0.1.4
[159] dqrng_0.4.1 spatstat.univar_3.1-2
[161] lazyeval_0.2.2 shiny_1.10.0
[163] htmltools_0.5.8.1 sctransform_0.4.1
[165] glue_1.8.0 tcltk_4.4.3
[167] spam_2.11-1 XVector_0.44.0
[169] RCurl_1.98-1.17 rprojroot_2.0.4
[171] scran_1.32.0 gridExtra_2.3
[173] sccore_1.0.5 R6_2.6.1
[175] tidyr_1.3.1 CompQuadForm_1.4.3
[177] labeling_0.4.3 cluster_2.1.8.1
[179] Rhdf5lib_1.26.0 DelayedArray_0.30.1
[181] tidyselect_1.2.1 vipor_0.4.7
[183] car_3.1-3 future_1.34.0
[185] leidenAlg_1.1.4 rsvd_1.0.5
[187] munsell_0.5.1 KernSmooth_2.23-26
[189] furrr_0.3.1 data.table_1.17.0
[191] htmlwidgets_1.6.4 RColorBrewer_1.1-3
[193] rlang_1.1.5 spatstat.sparse_3.1-0
[195] spatstat.explore_3.4-2 beeswarm_0.4.0